Welcome to DeLTA’s documentation!#
DeLTA (Deep Learning for Time-lapse Analysis) is a deep learning-based image processing pipeline for segmenting and tracking single cells in time-lapse microscopy movies.
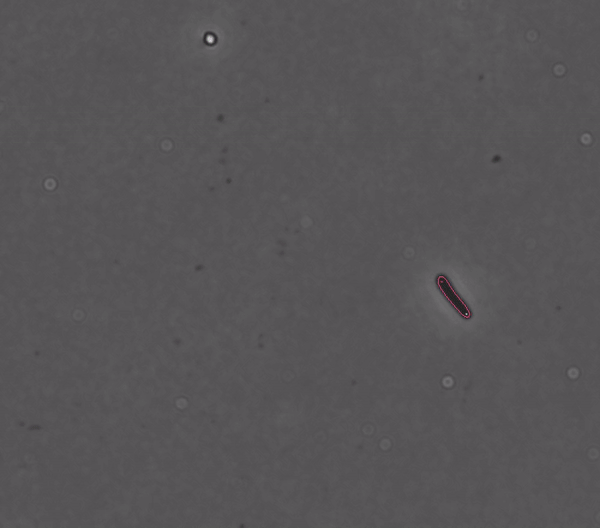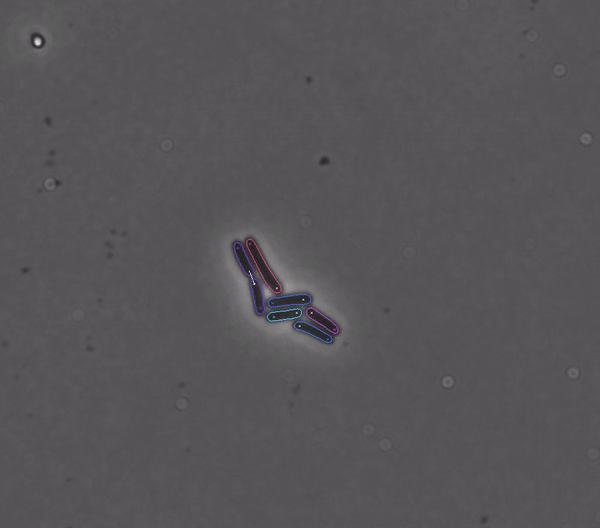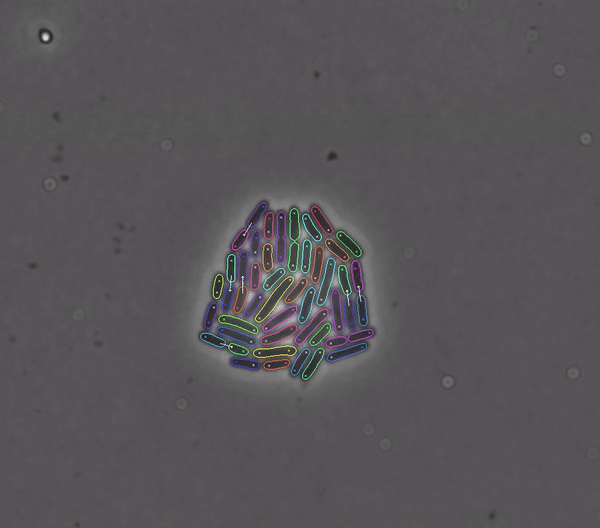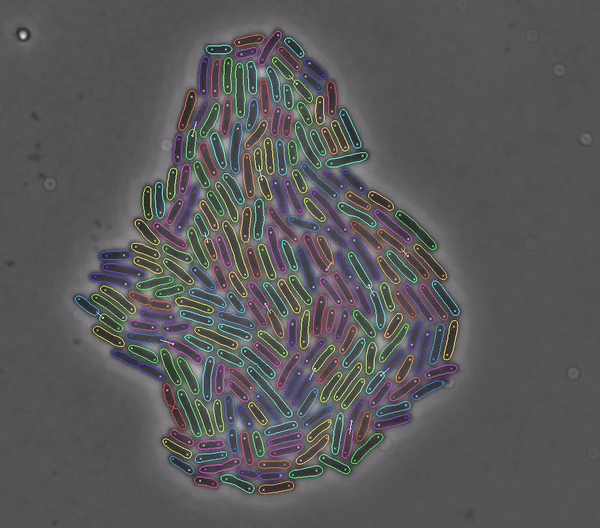
Getting started#
You can quickly check how DeLTA performs on your data for free by using our Google Colab notebook
To get started on your own system, check out the installation instructions, and run the pipeline on our data or your own.
See also our example scripts and results analysis examples.
Finally, you can contribute to DeLTA.
Gitlab repository: https://gitlab.com/delta-microscopy/delta
🐛 If you encounter bugs, have questions about the software, suggestions for new features, or even comments about the documentation, please use Gitlab’s issue system.
The main branch of the git repository is where new features and bug fixes
are integrated after being developed on feature branches. Stable releases
of DeLTA are tagged with git tags (and published on pypi or conda-forge).
Overview#
DeLTA revolves around a pipeline that can process movies of rod-shaped bacteria growing in 2D setups such as agarose pads, as well as movies of E. coli cells trapped in a microfluidic device known as a “mother machine”. Our pipeline is centered around two U-Net neural networks that are used sequentially:
To perform semantic binary segmentation of our cells as in the original U-Net paper.
To track cells from one movie frame to the next, and to identify cell divisions and mother/daughter cells.
A third U-Net can be used to identify regions of interest in the image before performing segmentation. This is used with mother machine movies to identify single chambers, but by default only 1 ROI covering the entire field-of-view is used in the 2D version. The U-Nets are implemented in Tensorflow 2 via the Keras API.
Citation#
If you use DeLTA in academic research, please cite us using one of the following references: